The Problem
Protein microarrays display proteins in high spatial density on a microscopic surface.
They can be used to test a variety of functions of many proteins simultaneously including interactions with other macromolecules, functional activity, and their suitability as substrates of enzymes.
Historically, these arrays have been produced by expressing and purifying proteins, which are then printed on the array surface. However, many challenges accompany this approach including the difficulty in purifying many proteins, a dynamic range in yields that spans several orders of magnitude (which is reflected in the widely varying amount of protein displayed on the array surface) and the danger of proteins becoming unfolded during the many manipulations, such as purification, storage and printing.
Our Solution
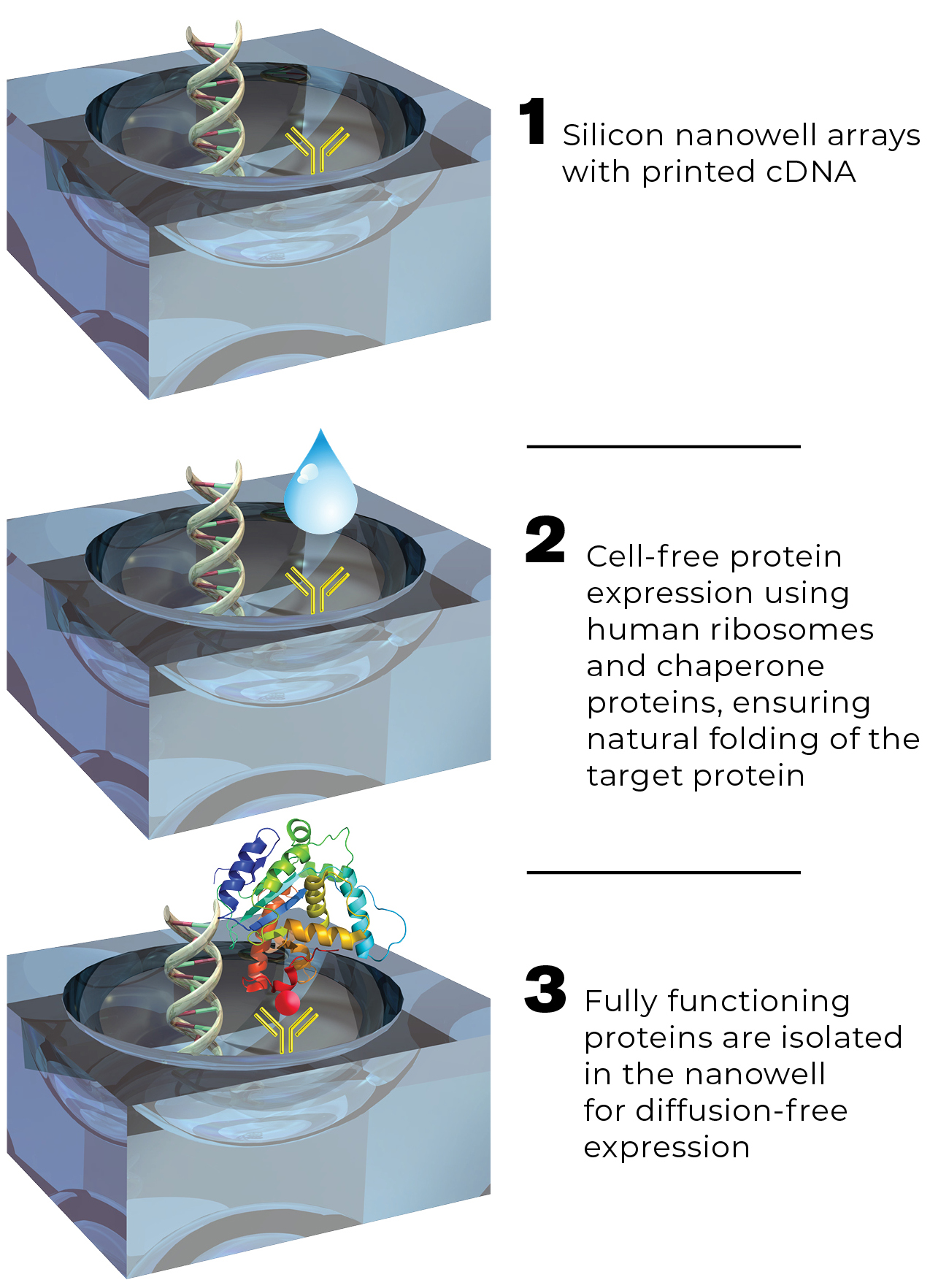
To avoid these challenges and to produce the most functional arrays possible, LaBaer and colleges developed a novel protein microarray technology, called Nucleic Acid-Programmable Protein Array (NAPPA), which replaces the complex process of spotting purified proteins with the simple process of spotting plasmid DNA. Our technology exploits the ability to transfer protein encoding regions into specialized tagged expression vectors. Proteins corresponding to the cDNAs are produced in the situ within the nanowell at translation (IVTT)-coupled cell lysates. The cDNAs are configured to append a common epitope tag to all of the proteins on their C-termini so that they can be captured by a high-affinity capture reagent that is immobilized along with the cDNA.
Advantages
This method minimizes direct manipulation of the proteins and produces them just-in-time for the experiment, avoiding problems with protein purification and stability (Science. 2004 305:86 and Nat Methods. 2008 5:535).
Replaces printing proteins with the more reliable and less expensive process of printing DNA.
Avoids the need to express, purify and store proteins
Displays better than 95% of sequence-verified full-length proteins
Display proteins at consistent levels 93% of proteins within two fold of the mean
Assures protein integrity by using both mammalian expression machinery and chaperone proteins to synthesize and fold proteins
Fully functioning proteins are isolated in the nanowell for diffusion-free expression
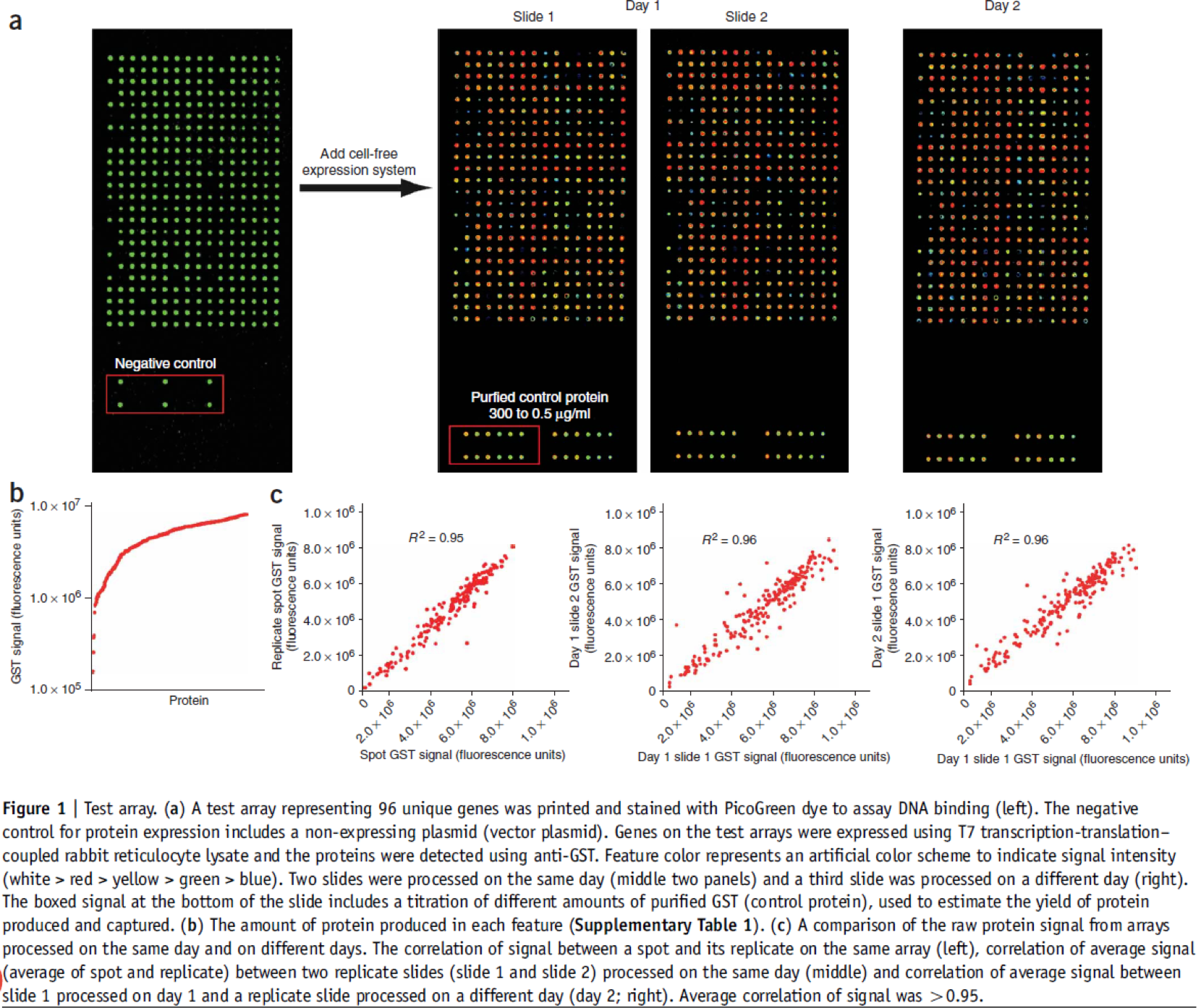
An example of a typical NAPPA array is shown below. DNA is visualized by PicoGreen and protein display by probing with an anti-GST antibody.
Protein Display Using Human Lysate